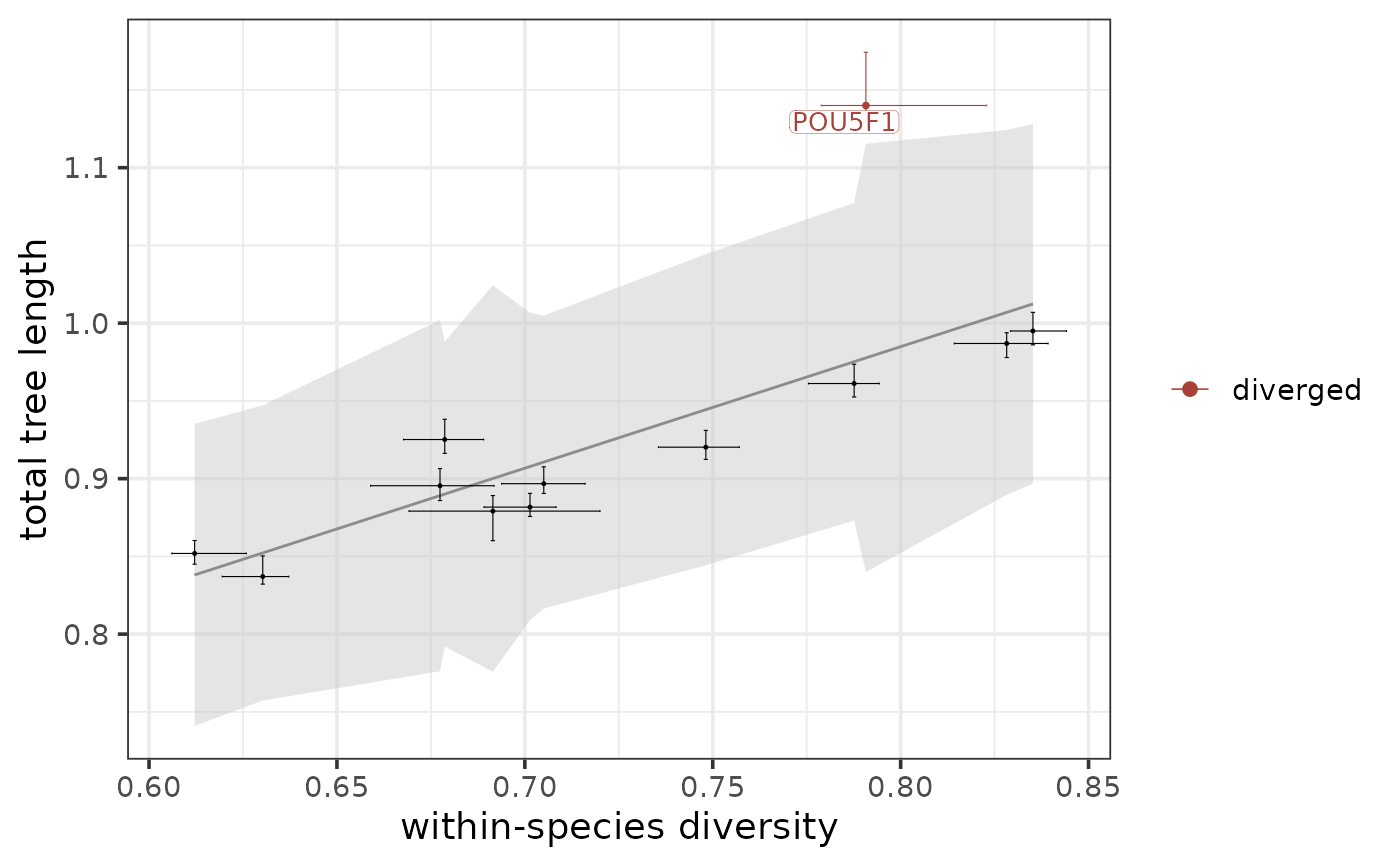
Plots the tree-bases statistics that characterize the cross-species conservation of the modules and marks the modules that were found to be conserved/diverged.
Usage
plotConservedDivergedModules(
module_conservation,
N = 5L,
rank_by = "residual",
colors = NULL,
font_size = 14,
label_size = 3.5
)Arguments
- module_conservation
Data frame of cross-species conservation measures per module.
Columns required in case the focus of interest is conservation and overall divergence:
- focus
Character, the focus of interest in terms of cross-species conservation, "overall" if the focus of interest is conservation and overall divergence.
- regulator
Character, transcriptional regulator.
- total_tree_length
Numeric, total tree length per module (typically the median across all jackknife versions of the module).
- lwr_total_tree_length
Numeric, the lower bound of the confidence interval of the total tree length calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- upr_total_tree_length
Numeric, the upper bound of the confidence interval of the total tree length calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- within_species_diversity
Numeric, within-species diveristy per module (typically the median across all jackknife versions of the module).
- lwr_within_species_diversity
Numeric, the lower bound of the confidence interval of the within-species diversity calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- upr_within_species_diversity
Numeric, the upper bound of the confidence interval of the within-species diversity calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- fit
Numeric, the fitted total tree length at the within-species diversity value of the module.
- lwr_fit
Numeric, the lower bound of the prediction interval of the fit.
- upr_fit
Numeric, the upper bound of the prediction interval of the fit.
- residual
Numeric, the residual of the module in the linear model. It is calculated as the difference between the observed and expected (fitted) total tree lengths.
- t_score
Numeric, the t-score of the module. It is calculated as the residual normalized by the standard error of the total tree length prediction at the given within-species diversity value
- conservation
Character, "not_significant" if the module falls inside the prediction interval of the fit, "diverged" if a module has a higher total tree length than the upper boundary of the prediction interval, and "conserved" if a module has a lower total tree length than the lower boundary of the prediction interval
Columns required in case the focus of interest is species-specific divergence:
- focus
Character, the focus of interest in terms of cross-species conservation, the name of a species if the focus of interest is species-specific divergence
- regulator
Character, transcriptional regulator.
- {{species}}_subtree_length
Numeric, the subtree length of the species per module (typically the median across all jackknife versions of the module).
- lwr_{{species}}_subtree_length
Numeric, the lower bound of the confidence interval of the species subtree length calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- upr_{{species}}_subtree_length
Numeric, the upper bound of the confidence interval of the species subtree length calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- {{species}}_diversity
Numeric, the diveristy of the species of interest per module (typically the median across all jackknife versions of the module).
- lwr_{{species}}_diversity
Numeric, the lower bound of the confidence interval of the species-specific diversity calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- upr_{{species}}_diversity
Numeric, the upper bound of the confidence interval of the species-specific diversity calculated based on the jackknifed versions of the module (optional, only needed for plotting error bars later on).
- fit
Numeric, the fitted subtree length of a species.
- lwr_fit
Numeric, the lower bound of the prediction interval of the fit.
- upr_fit
Numeric, the upper bound of the prediction interval of the fit.
- residual
Numeric, the residual of the module in the linear model. It is calculated as the difference between the observed and expected (fitted) subtree length of a species.
- t_score
Numeric, the t-score of the module. It is calculated as the residual normalized by the standard error of the species subtree length prediction.
- conservation
Character, 'not_significant' if the module falls inside the prediction interval of the fit, 'diverged' if a module has a higher species subtree length than the upper boundary of the prediction interval, and 'conserved' if a module has a lower species subtree length than the lower boundary of the prediction interval.
- N
Integer, the number of top conserved and diverged modules to label.
- rank_by
Character, one of "residual" and "t_score". The name of the variable to rank the by when selecting the top N conserved and diverged modules to label.
- colors
(Named) character vector of length 2, the colors for the diverged and conserved modules.
- font_size
Numeric, font size (default: 14).
- label_size
Numeric, the size of the labels for the most conserved and diverged modules (default: 3.5).
Details
To determine whether a module as whole is conserved, diverged overall or diverged on a specific lineage, the CroCoNet approach relies on module trees reconstructed from pairwise preservation scores between replicates and statistics calculated based on these trees (total tree length, subtree lengths of the species, overall within-species and species-specific diversity, for details please see calculatePresStats, reconstructTrees, calculateTreeStats and findConservedDivergedModules).
The function plots each input module along the regression line that captures the relationship between total tree length and within-species diversity (in case the focus of interest is conservation and overall divergence) or between the subtree length and diversity of a species (in case the focus of interest is divergence between this particular species and all others). A module is considered diverged overall/in a species-specific manner if it falls above the prediction interval of the regression line, while a module is considered conserved if it falls below the prediction interval of the regression line.
The regression line is drawn in dark grey, and the 95% prediction interval of the line is shown as a light grey area. If the focus of interest is conservation and overall divergence, the conserved modules are colored green, the diverged modules are colored red, and the top 'N' conserved and diverged modules based on the variable specified in rank_by are labelled. If the focus of interest is divergence between a particular species and all others, only the diverged modules are colored and labelled, the modules below the prediction interval are not meaningful and thus not highlighted. If confidence intervals are provided for the variables, each module is plotted with error bars.