CroCoNet is a framework to quantitatively compare gene regulatory networks across species and identify conserved and diverged modules. It hinges on contrasting module variability within and across species in order to distinguish true evolutionary divergence from confounding factors such as diversity across individuals, environmental differences and technical noise.
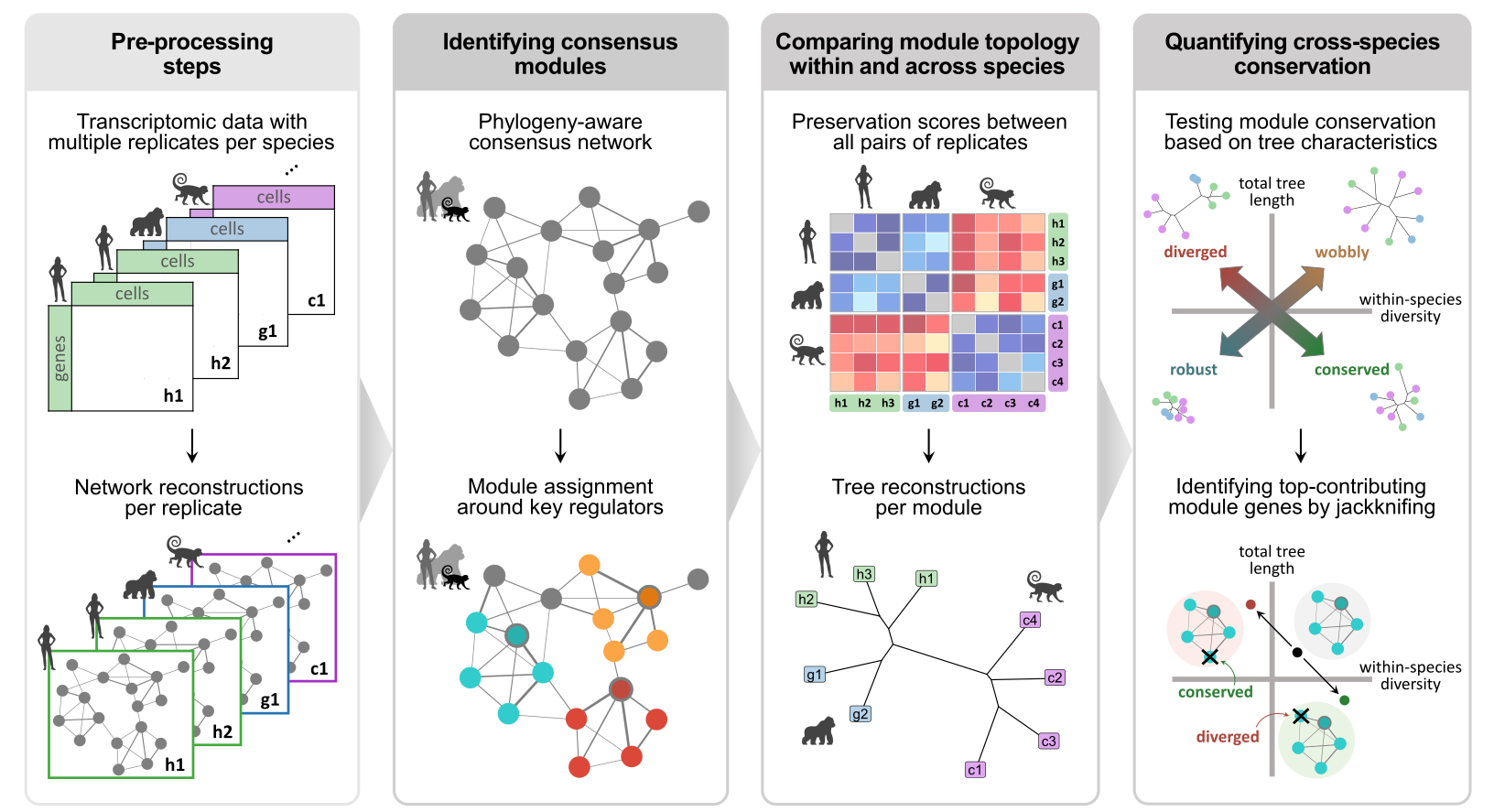
The main steps of the CroCoNet workflow
⬇️ Installation
For the installation, the R package devtools is needed.
install.packages("devtools")
library(devtools)Once devtools is available, you can install the development version of CroCoNet and all its dependencies from GitHub with:
devtools::install_github("Hellmann-Lab/CroCoNet")📖 User guide
For a step-by-step guide and detailed explanations, please check out the vignette on the analysis of an example scRNA-seq dataset:
browseVignettes("CroCoNet")You can access this vignette, along with the documentation of all functions, on the CroCoNet website as well.
